ChronoRoot: High-throughput phenotyping by deep segmentation networks reveals novel temporal parameters of plant root system architecture
Abstract
Background
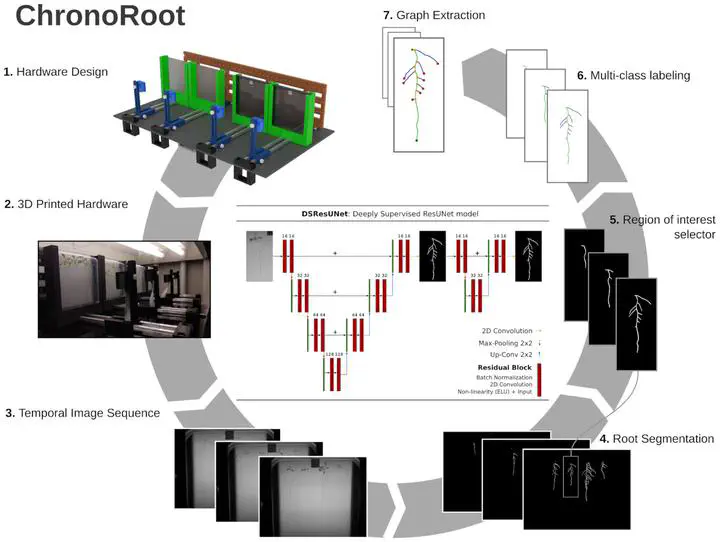
Deep learning methods have outperformed previous techniques in most computer vision tasks, including image-based plant phenotyping. However, massive data collection of root traits and the development of associated artificial intelligence approaches have been hampered by the inaccessibility of the rhizosphere. Here we present ChronoRoot, a system that combines 3D-printed open-hardware with deep segmentation networks for high temporal resolution phenotyping of plant roots in agarized medium.Results
We developed a novel deep learning-based root extraction method that leverages the latest advances in convolutional neural networks for image segmentation and incorporates temporal consistency into the root system architecture reconstruction process. Automatic extraction of phenotypic parameters from sequences of images allowed a comprehensive characterization of the root system growth dynamics. Furthermore, novel time-associated parameters emerged from the analysis of spectral features derived from temporal signals.Conclusions
Our work shows that the combination of machine intelligence methods and a 3D-printed device expands the possibilities of root high-throughput phenotyping for genetics and natural variation studies, as well as the screening of clock-related mutants, revealing novel root traits.Type
Publication
GigaScience